柱状图和箱线图的代码能理解了其实发现好多作图都是可以触类旁通的,小提琴图作为科研结果常用展示图也不可或缺,用ggplot或者vioplot。
library("vioplot")
#准备数据
ToothGrowth$dose <- as.factor(ToothGrowth$dose)
p <- ggplot(ToothGrowth, aes(x=dose, y=len)) +
geom_violin(trim=T) #T改为F即完整的小提琴图
p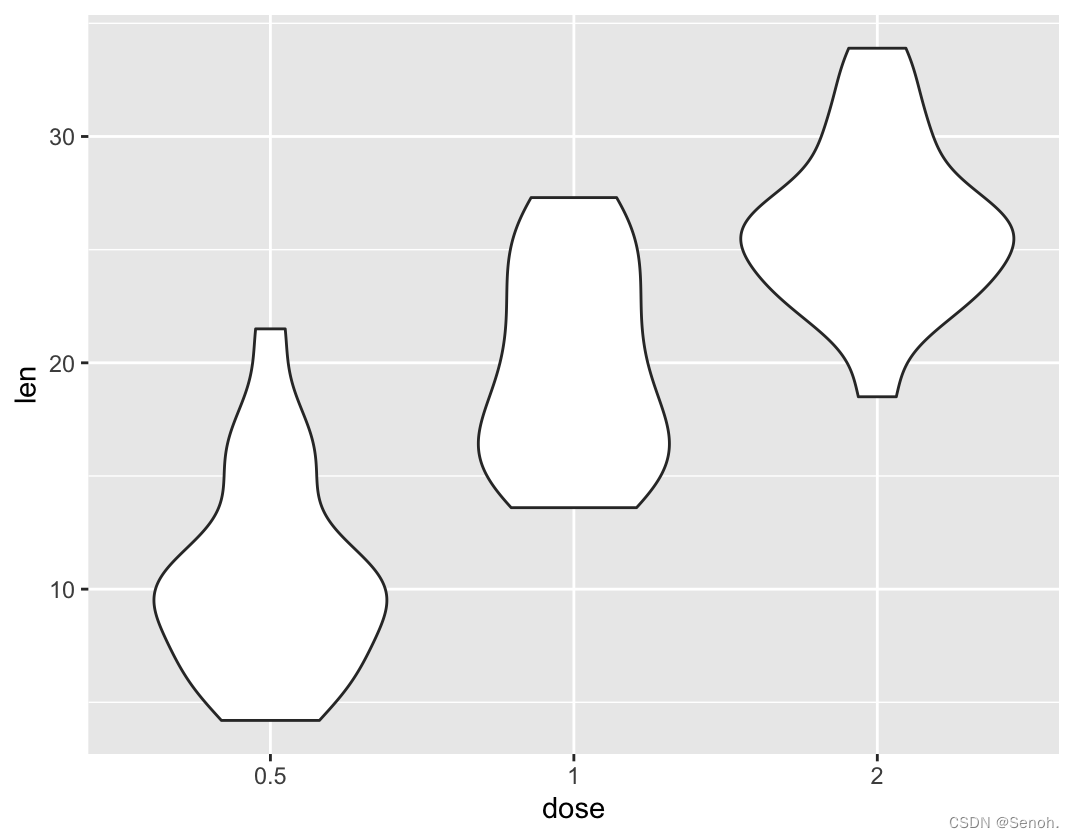
#选择需要显示的x项
p + scale_x_discrete(limits=c("0.5", "2"))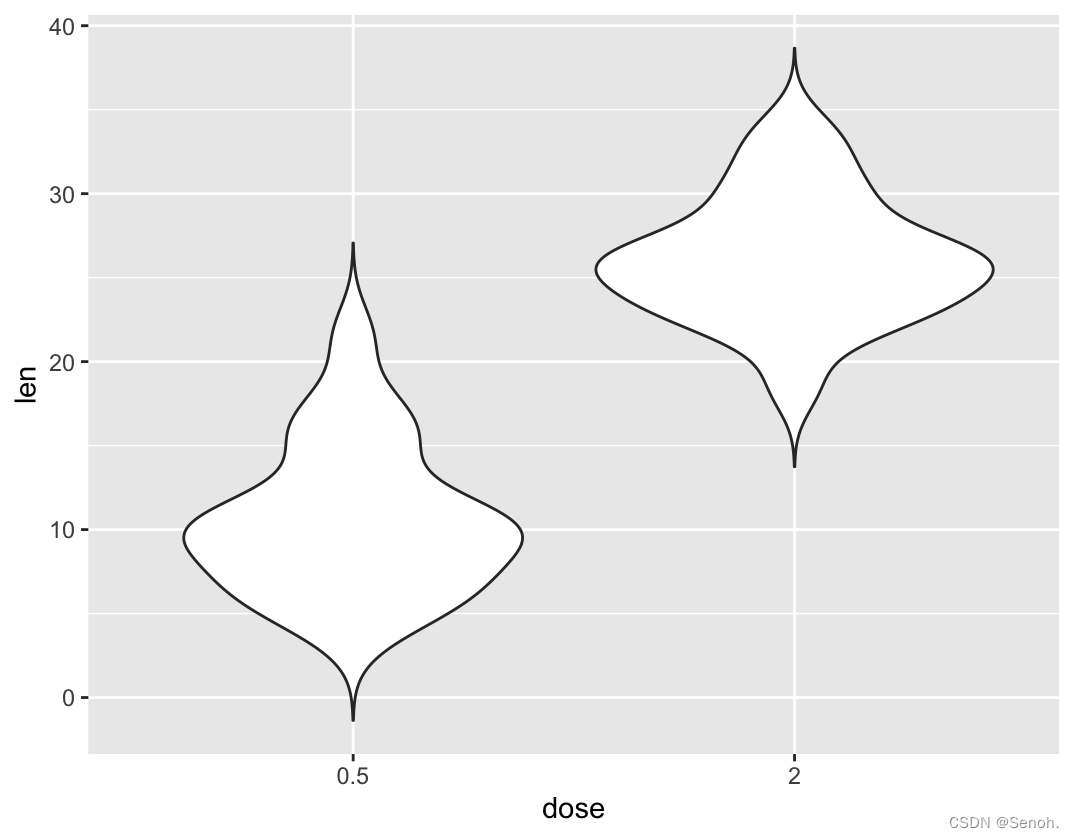
#添加中位值median,均值改mean
p + stat_summary(fun.y=mean, geom="point", shape=23, size=2, color="red")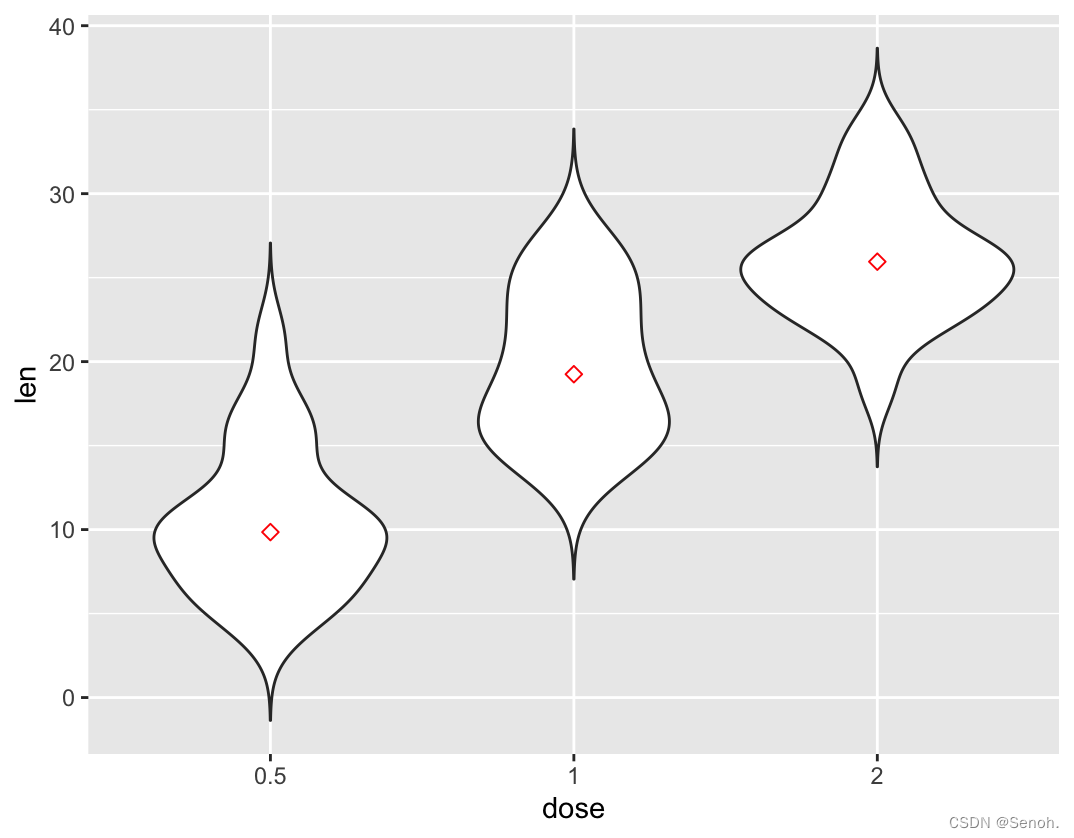
#中位数和四分位数
p + geom_boxplot(width=0.1)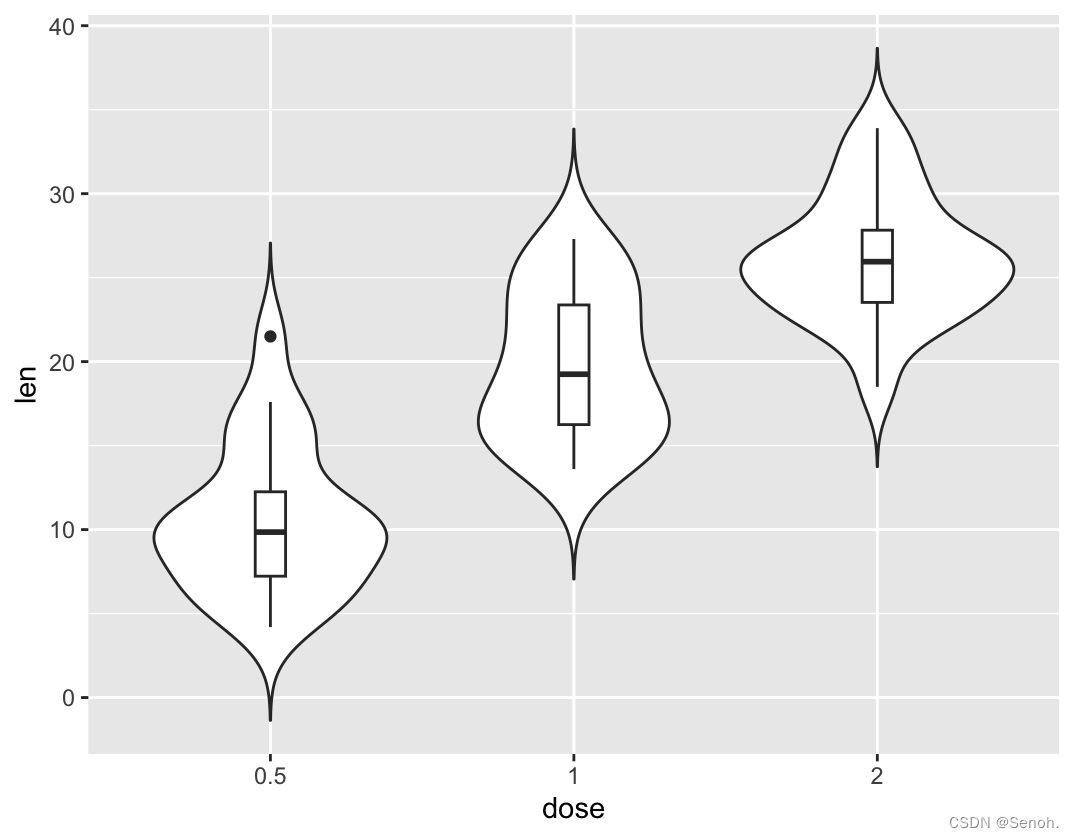
#均值和标准差
p <- ggplot(ToothGrowth, aes(x=dose, y=len)) +
geom_violin(trim=FALSE)
p + stat_summary(fun.data=mean_sdl, mult=2,
geom="pointrange", color="red")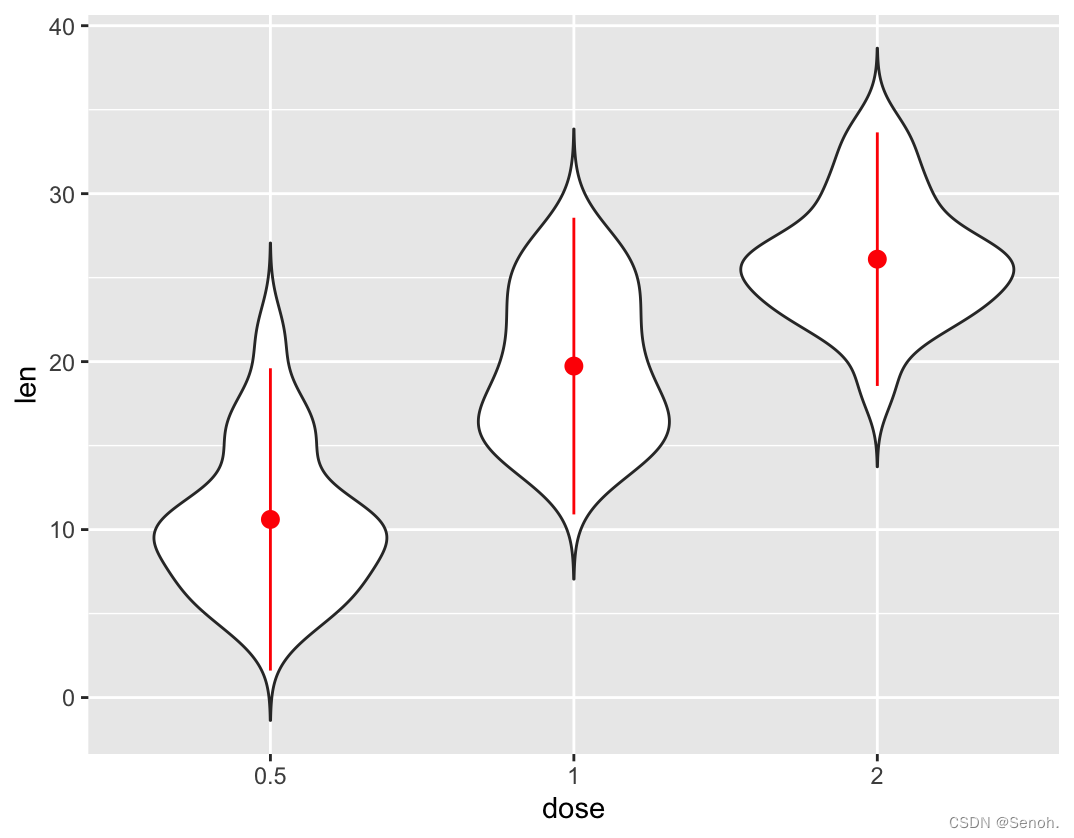
#带点小提琴图
p + geom_dotplot(binaxis='y', stackdir='center', dotsize=1)
#点发散
p + geom_jitter(shape=16, position=position_jitter(0.2))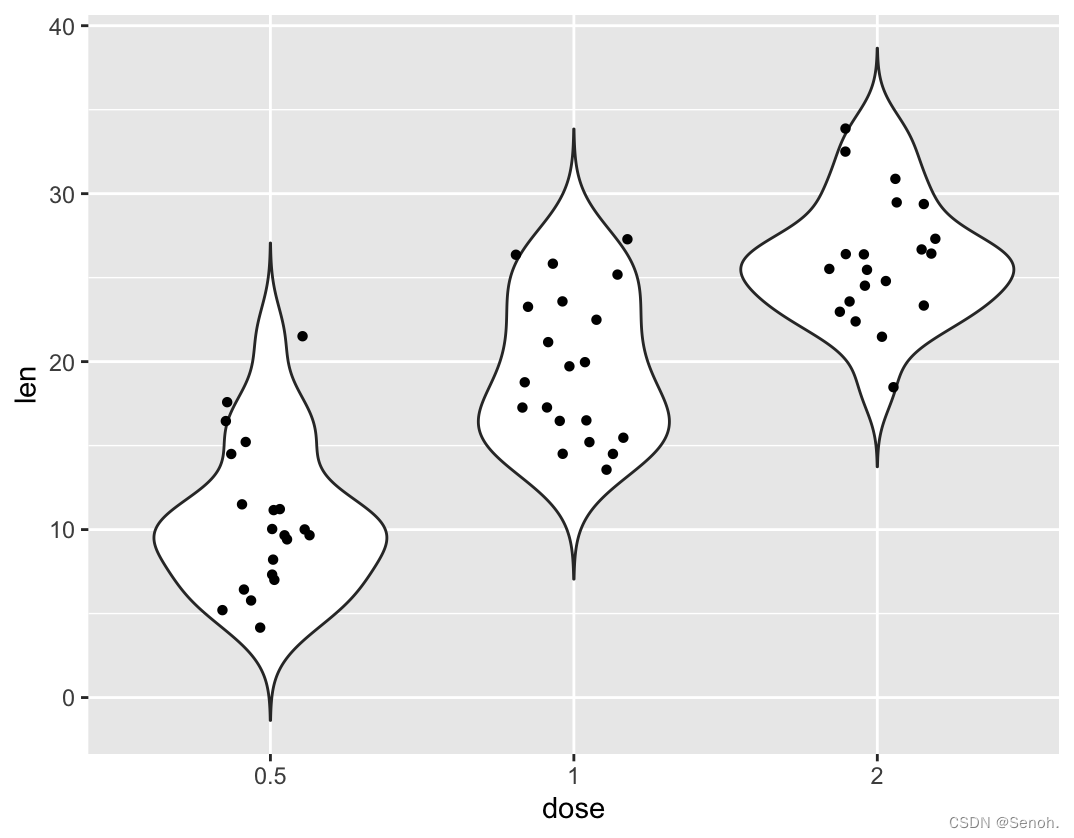
定义颜色的代码跟前两章柱状图箱线图的一样,都对应着线框颜色和填充颜色。
#准备数据
set.seed(1)
n <- 10000
ii <- rbinom(n, 1, 0.5)
data <- rnorm(n, mean = 130, sd = 10) * ii +
rnorm(n, mean = 80, sd = 5) * (1 - ii)
#柱状图
hist(data, probability = TRUE,
col = "grey", axes = FALSE,
main = "", xlab = "", ylab = "")
axis(1)
#密度线
lines(density(data), lwd = 2, col = "red")
#添加小提琴图
par(new = TRUE)
vioplot(data, horizontal = TRUE,
yaxt = "n", axes = FALSE,
col = ggsci::pal_npg("nrc",alpha = 0.2)(1)) 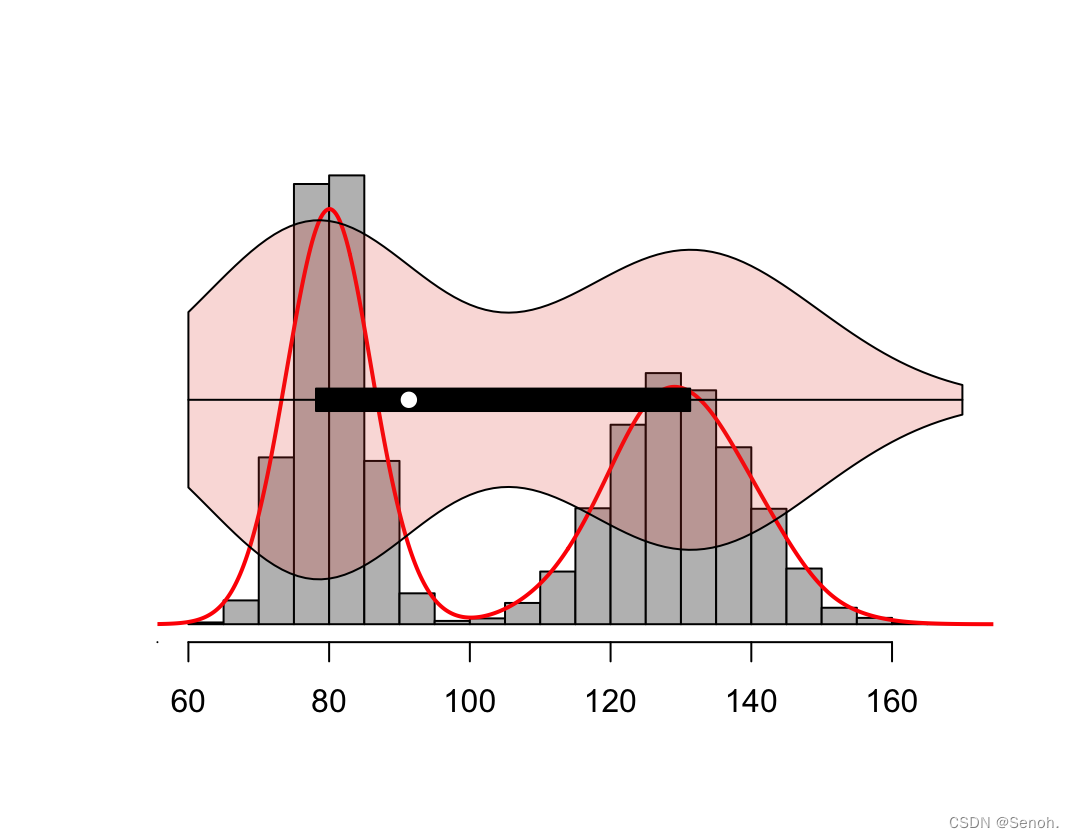
#准备数据,分组
data <- trees
tall <- trees[trees$Height >= 76, ]
small <- trees[trees$Height < 76, ]
#绘图
vioplot(tall, side = "left", plotCentre = "line", col = "#3B4992B2")
vioplot(small, side = "right", plotCentre = "line", col = "#EE0000B2", add = TRUE)
#图例
legend("topleft", legend = c("Tall", "Small"), fill = c("#3B4992B2","#EE0000B2"), cex = 1.25)
#分画布
par(mfrow = c(1, 2))
data <- chickwts
#低到高
medians <- reorder(data$feed, data$weight, median)
#medians <- with(data, reorder(feed, weight, median)) # 也可用这行,效果一样的,下面同理
vioplot(data$weight ~ medians, col = 2:(length(levels(data$feed)) + 1),
xlab = "", ylab = "Weight", las = 2)
#高到低
medians <- reorder(data$feed, -data$weight, median)
vioplot(data$weight ~ medians, col = 2:(length(levels(data$feed)) + 1),
xlab = "", ylab = "Weight", las = 2)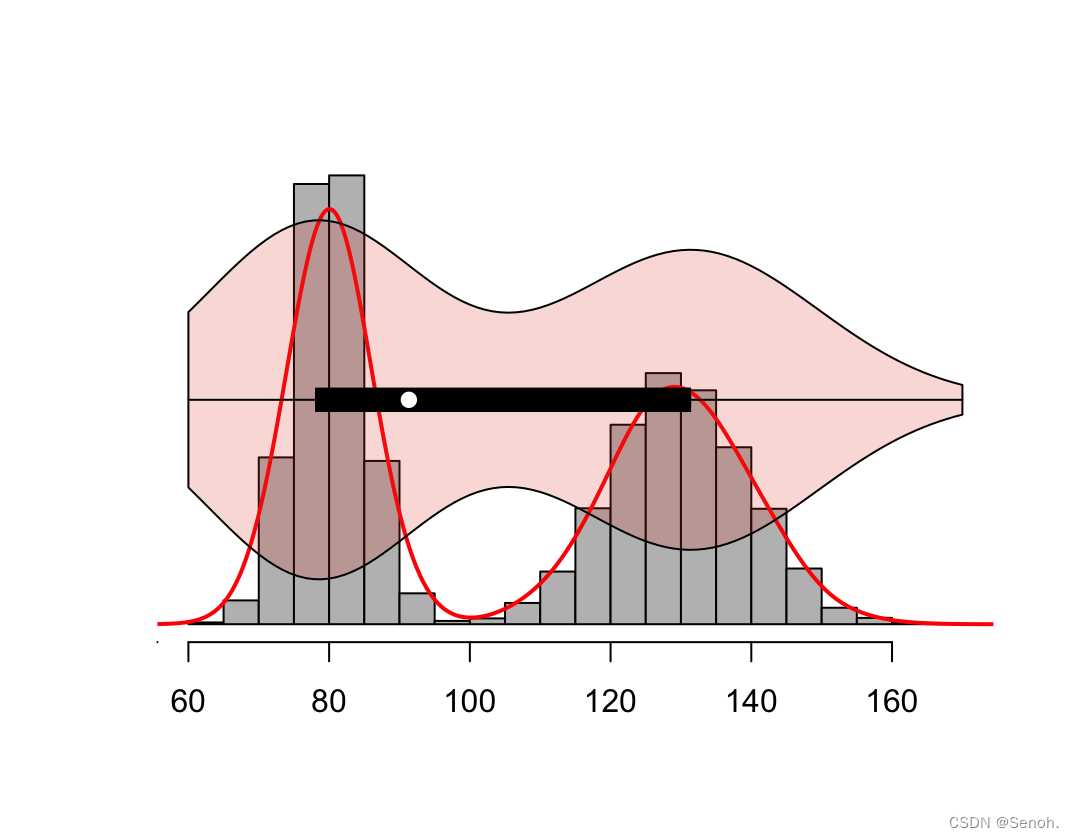
library("ggpubr")
library("reshape2")
norcol="#EE0000FF" #疾病组
concol="#3B4992FF" #正常组
#读取文件
rt=read.table(inputFile,header=T,sep="\t",check.names=F,row.names=1)
x=colnames(rt)[1]
colnames(rt)[1]="Type"
#转换分类矩阵数据
data=melt(rt,id.vars=c("Type"))
colnames(data)=c("Type","Gene","Expression")
#绘图
pdf(file=outFile, width=6, height=5)
p=ggviolin(data, x="Gene", y="Expression", color = "Type",
ylab="Gene expression",
xlab="Gene",
#legend.title=x,
add.params = list(fill="white"),
palette = c(concol,norcol),
width=1, add = "boxplot")
#p=p+rotate_x_text(60)
p1=p+stat_compare_means(aes(group=Type),
method="wilcox.test", #可换其他统计方法
symnum.args=list(cutpoints = c(0, 0.001, 0.01, 0.05, 1), symbols = c("***", "**", "*", " ")),
label = "p.signif")
print(p1)
dev.off()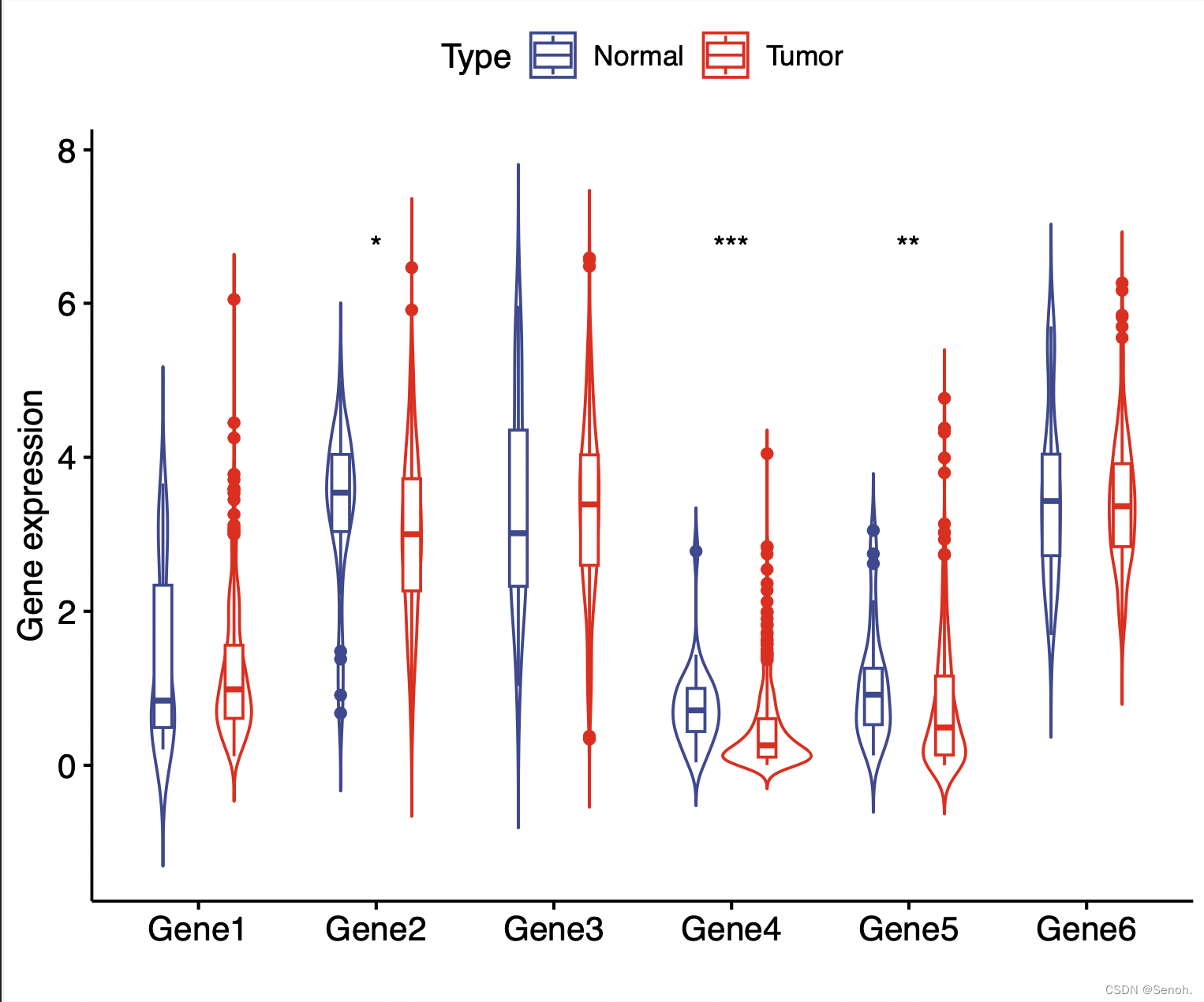
library(ggpubr)
library(ggsci)
ggsci::pal_tron("legacy")(4)
color=c("#FF410DFF","#6EE2FFFF","#F7C530FF","#95CC5EFF")
#读取文件
rt=read.table(inputFile,header=T,sep="\t",check.names=F)
x=colnames(rt)[2]
y=colnames(rt)[3]
colnames(rt)=c("id","Type","Expression")
#设置分组
group=levels(factor(rt$Type))
rt$Type=factor(rt$Type, levels=group)
comp=combn(group,2)
my_comparisons=list()
for(i in 1:ncol(comp)){my_comparisons[[i]]<-comp[,i]}
#绘图
pdf(file=outFile, width=6, height=5)
ggviolin(rt, x="Type", y="Expression", fill = "Type",
xlab=x, ylab=paste0(y," expression"),
legend.title=x,
palette = color,
add = "boxplot", add.params = list(fill="white"))+
#stat_compare_means(comparisons = my_comparisons) #显示具体数值
stat_compare_means(comparisons = my_comparisons,
symnum.args=list(cutpoints = c(0, 0.001, 0.01, 0.05, 1),
symbols = c("***", "**", "*", "ns")),
label = "p.signif") #显著替换成星号
dev.off()
library(ggplot2)
library(reshape2)
color = c("#FF410DFF","#6EE2FFFF","#F7C530FF","#95CC5EFF")
#读取文件
rt=read.table(inputFile, header=T,sep="\t",check.names=F,row.names=1)
x=colnames(rt)[1]
colnames(rt)[1]="Type"
#计算组间差异
geneSig=c("")
for(gene in colnames(rt)[2:ncol(rt)]){
rt1=rt[,c(gene,"Type")]
colnames(rt1)=c("expression","Type")
p=1
if(length(levels(factor(rt1$Type)))>2){
test=kruskal.test(expression ~ Type, data = rt1)
p=test$p.value
}else{
test=wilcox.test(expression ~ Type, data = rt1)
p=test$p.value
}
Sig=ifelse(p<0.001,"***",ifelse(p<0.01,"**",ifelse(p<0.05,"*","")))
geneSig=c(geneSig,Sig)# 根据p值显著标星号
}
colnames(rt)=paste0(colnames(rt),geneSig)
#转换数据类型
data=melt(rt,id.vars=c("Type"))
colnames(data)=c("Type","Gene","Expression")
#绘图
pdf(file=outFile, width=9, height=5)
p1=ggplot(data,aes(x=Type,
y=Expression,
fill=Type))+
guides(fill=guide_legend(title=x))+
labs(x = x, y = "Gene expression")+ #y轴
geom_violin()+
scale_fill_manual(values = color) + geom_boxplot(width=0.2,position=position_dodge(0.9))+
facet_wrap(~Gene,nrow =1)+
theme_bw()+
theme(axis.text.x = element_text(angle = 45, hjust = 1))
print(p1)
dev.off()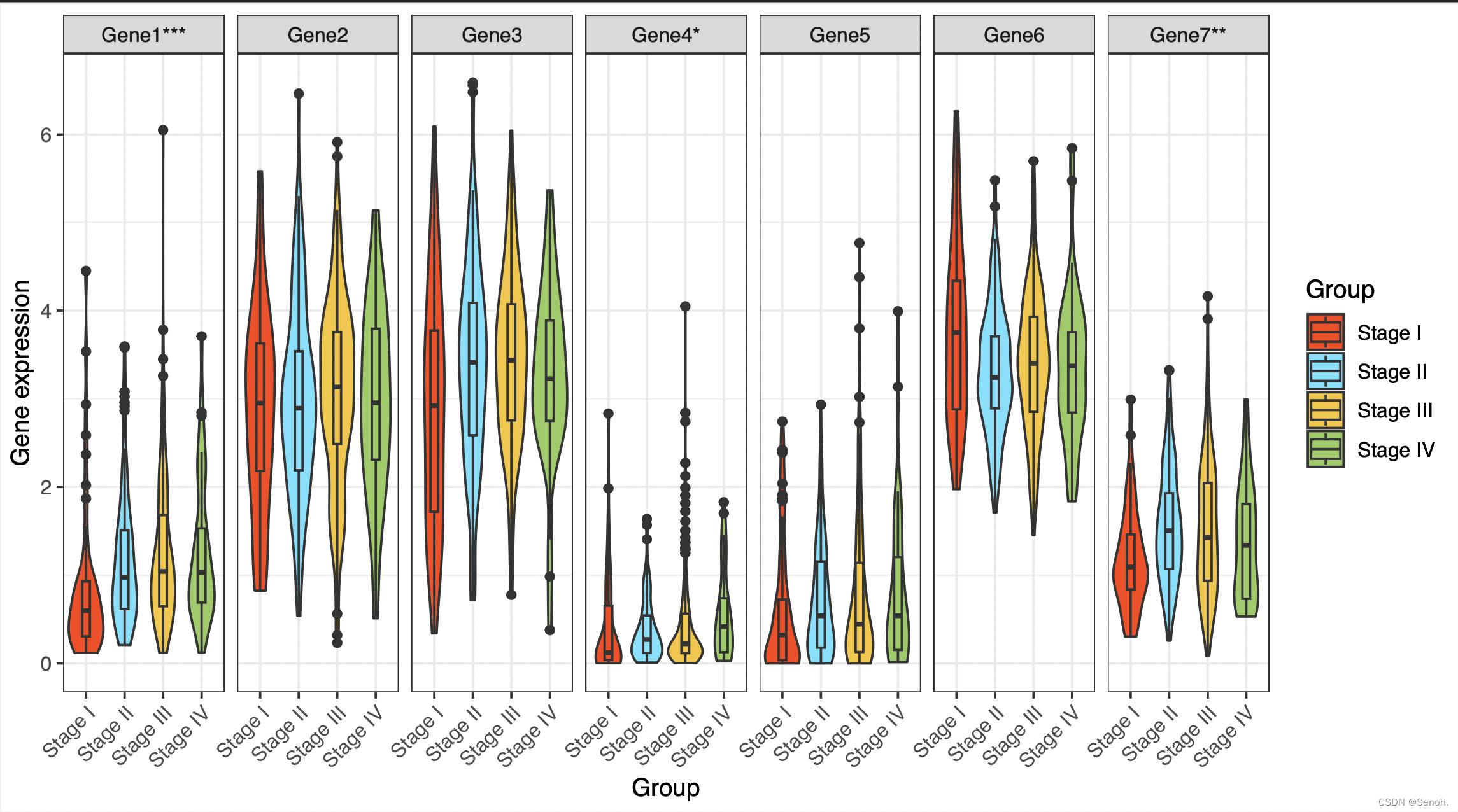
小提琴图和箱线图其实大同小异,都是常用于展示基因组间差异表达,很基础的描述性统计可视化图。和violinplot区分开来,部分代码不同,但是效果和目的一样。每个图都可以自己换线框、填充颜色透明度等,发文不重复。